Freesurfer Subparcellation: Difference between revisions
No edit summary |
|||
| (9 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
Freesurfer annotations and labels describe swaths of cortical regions, defined anatomically during the recon-all process, or functionally or statistically (e.g., significant GLM clusters). Tools exist to further subdivide these regions to the desired level of granularity. | Freesurfer annotations and labels describe swaths of cortical regions, defined anatomically during the recon-all process, or functionally or statistically (e.g., significant GLM clusters). Tools exist to further subdivide these regions to the desired level of granularity. | ||
= Using AnchorBar = | |||
AnchorBar is a set of Python tools developed in our lab that maintain a database of a set of annotations and supports set operations on the labels associated with one or more annotations. For example, an atlas-based annotation can be intersected with a functional annotation (e.g., from a group-level GLM) to subdivide large functional clusters along anatomical boundaries. | |||
= Using mris_divide_parcellation = | |||
This program divides one or more parcellations into divisions | This program divides one or more parcellations into divisions | ||
perpendicular to the long axis of the label. The number of divisions | perpendicular to the long axis of the label. The number of divisions | ||
| Line 34: | Line 36: | ||
divisions. The first division has the same name as the original label. | divisions. The first division has the same name as the original label. | ||
== Sample Split File (Functional Clusters) == | |||
In this example, I have carried out a one-sample group mean (OSGM) analysis and did a cluster-size threshold. This saved a number of clusters in two .annot files (one in each hemisphere). There are also some ''*.cluster.summary'' files in this directory. Reading these files, I see the smallest clusters are about 120mm<sup>2</sup>. I would like to subdivide the larger clusters so that they are all between about 100-150mm<sup>2</sup>. I load the corresponding .annot file in Freesurfer just to verify the cluster names (the summary file lists 10 clusters, but the names do not follow a consistent naming convention). From the file, I see that clusters 1,2,3,4,6 and 8 need to be subdivided. Simple division by the desired cluster size tells me the number of divisions I want for each cluster: | In this example, I have carried out a one-sample group mean (OSGM) analysis and did a cluster-size threshold. This saved a number of clusters in two .annot files (one in each hemisphere). There are also some ''*.cluster.summary'' files in this directory. Reading these files, I see the smallest clusters are about 120mm<sup>2</sup>. I would like to subdivide the larger clusters so that they are all between about 100-150mm<sup>2</sup>. I load the corresponding .annot file in Freesurfer just to verify the cluster names (the summary file lists 10 clusters, but the names do not follow a consistent naming convention). From the file, I see that clusters 1,2,3,4,6 and 8 need to be subdivided. Simple division by the desired cluster size tells me the number of divisions I want for each cluster: | ||
| Line 75: | Line 77: | ||
Note: the first time I ran this, I have no idea where the rh.functional_subclusters.annot file went! Running the lh and re-running the rh subparcellation generated the expected files in the fsaverage/label directory. | Note: the first time I ran this, I have no idea where the rh.functional_subclusters.annot file went! Running the lh and re-running the rh subparcellation generated the expected files in the fsaverage/label directory. | ||
== Check Subparcellation == | |||
Presumably the python wrapper does this iteratively because all subdivisions are done along the same axis. As I discovered, a large number of subdivisions may be realized as a series of thin bands (below). In this case, it might be best to subdivide larger regions over a series of calls. | Presumably the python wrapper does this iteratively because all subdivisions are done along the same axis. As I discovered, a large number of subdivisions may be realized as a series of thin bands (below). In this case, it might be best to subdivide larger regions over a series of calls, as I describe below. | ||
[[File:Striped_Parcellation.png]] | [[File:Striped_Parcellation.png]] | ||
| Line 94: | Line 96: | ||
200 200functional_subclusters | 200 200functional_subclusters | ||
Because each call to <code>mris_divide_parcellation</code> bisects the region along it's longest axis, successive function calls at each iteration produce increasingly less elongated subparcellations. | |||
== Transfer Subparcellation to Other Subjects == | == Transfer Subparcellation to Other Subjects == | ||
Once satisfied with the source subparcellation, the scheme can be mapped to other surfaces using mri_surf2surf (this needs to be done individually for the lh and rh surfaces -- below is an example command for the left): | Once satisfied with the source subparcellation, the scheme ''can'' be mapped to other surfaces using mri_surf2surf if, for example, you are working in '''self''' space (this needs to be done individually for the lh and rh surfaces -- below is an example command for the left): | ||
mri_surf2surf \ | mri_surf2surf \ | ||
| Line 105: | Line 107: | ||
--sval-annot $SUBJECTS_DIR/fsaverage/label/lh.200functional_subclusters.annot \ | --sval-annot $SUBJECTS_DIR/fsaverage/label/lh.200functional_subclusters.annot \ | ||
--tval $SUBJECTS_DIR/FS_T1_501/label/lh.200functional_subclusters.annot | --tval $SUBJECTS_DIR/FS_T1_501/label/lh.200functional_subclusters.annot | ||
''Pro Tip: Use a global variable (e.g. SUBN) and use $SUBN in place of your subject in trgsubject and tval. Better yet, use the clustmap.sh shell script.'' | |||
You may want to pull out overlapping voxels for T1 and T2 data (like a venn diagram) before mapping the surfaces to each subject, [http://ccnlab.psy.buffalo.edu/wiki/index.php/Working_with_ROIs_(Freesurfer) here]. | You may want to pull out overlapping voxels for T1 and T2 data (like a venn diagram) before mapping the surfaces to each subject, [http://ccnlab.psy.buffalo.edu/wiki/index.php/Working_with_ROIs_(Freesurfer) here]. | ||
'''Note: If you are working in ''fsaverage'' space for all participants, you only have to subparcellate your annotations once -- for the fsaverage surface.''' Because the functional analyses are invariably carried out in fsaverage space in order to compute group-level statistics, you will seldom have to transfer a subparcellation to a specific participant's surface space. | |||
[[Category: FreeSurfer]] | |||
[[Category: Network Analyses]] | [[Category: Network Analyses]] | ||
Latest revision as of 13:30, 27 September 2022
Freesurfer annotations and labels describe swaths of cortical regions, defined anatomically during the recon-all process, or functionally or statistically (e.g., significant GLM clusters). Tools exist to further subdivide these regions to the desired level of granularity.
Using AnchorBar
AnchorBar is a set of Python tools developed in our lab that maintain a database of a set of annotations and supports set operations on the labels associated with one or more annotations. For example, an atlas-based annotation can be intersected with a functional annotation (e.g., from a group-level GLM) to subdivide large functional clusters along anatomical boundaries.
Using mris_divide_parcellation
This program divides one or more parcellations into divisions perpendicular to the long axis of the label. The number of divisions can be specified in one of two ways, depending upon the nature of the fourth argument.
First, a splitfile can be specified as the fourth argument. The splitfile is a text file with two columns. The first column is the name of the label in the source annotation, and the second column is the number of units that label should be divided into. The names of the labels depends upon the source parcellation. For aparc.annot and aparc.a2005.annot, the names can be found in $FREESURFER_HOME/FreeSurferColorLUT.txt. For aparc.annot, the labels are between the ranges of 1000-1034. For aparc.a2005s.annot, the labels are between the ranges of 1100-1181. The name for the label is the name of the segmentation without the 'ctx-lh'. Note that the name included in the splitfile does not indicate the hemisphere. For example, 1023 is 'ctx-lh-posteriorcingulate'. You should put 'posteriorcingulate' in the splitfile. Eg, to divide it into three segments, the following line should appear in the splitfile:
posteriorcingulate 3
Only labels that should be split need be specified in the splitfile.
The second method is to specify an area threshold (in mm^2) as the fourth argument, in which case each label is divided until each subdivision is smaller than the area threshold.
The output label name will be the original name with _divN appended, where N is the division number. N will go from 2 to the number of divisions. The first division has the same name as the original label.
Sample Split File (Functional Clusters)
In this example, I have carried out a one-sample group mean (OSGM) analysis and did a cluster-size threshold. This saved a number of clusters in two .annot files (one in each hemisphere). There are also some *.cluster.summary files in this directory. Reading these files, I see the smallest clusters are about 120mm2. I would like to subdivide the larger clusters so that they are all between about 100-150mm2. I load the corresponding .annot file in Freesurfer just to verify the cluster names (the summary file lists 10 clusters, but the names do not follow a consistent naming convention). From the file, I see that clusters 1,2,3,4,6 and 8 need to be subdivided. Simple division by the desired cluster size tells me the number of divisions I want for each cluster:
echo cluster-001 6 > rsplittable.txt echo cluster-002 7 >> rsplittable.txt echo cluster-003 5 >> rsplittable.txt echo cluster-004 5 >> rsplittable.txt echo cluster-006 2 >> rsplittable.txt echo cluster-008 2 >> rsplittable.txt
Repeat the process for the lh clusters, creating an lsplittable.txt file.
The group analysis used the fsaverage surface. Since mris_divide_parcellation assumes we're subdividing a particular subject's annotation, it makes sense to copy the source .annot files into the fsaverage label/ folder (giving the appropriate ?h prefix)
cp cache.th30.abs.sig.ocn.annot \ $SUBJECTS_DIR\fsaverage\label\rh.cache.th30.abs.sig.ocn.annot
Then call the program thus (assuming that rsplittable.txt is in your current working directory):
mris_divide_parcellation fsaverage rh \ cache.th30.abs.sig.ocn.annot rsplittable.txt \ functional_subclusters
Program output:
reading colortable from annotation file... colortable with 11 entries read (originally none) interpreting 4th command line arg as split file name dividing cluster-001 (1) into 6 parts dividing cluster-002 (2) into 7 parts dividing cluster-003 (3) into 5 parts dividing cluster-004 (4) into 5 parts dividing cluster-006 (6) into 2 parts dividing cluster-008 (8) into 2 parts allocating new colortable with 31 additional units... saving annotation to functional_subclusters
Repeat for lh.
Note: the first time I ran this, I have no idea where the rh.functional_subclusters.annot file went! Running the lh and re-running the rh subparcellation generated the expected files in the fsaverage/label directory.
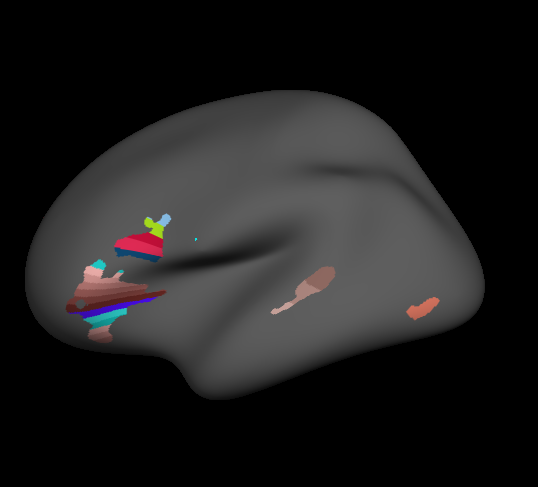
Check Subparcellation
Presumably the python wrapper does this iteratively because all subdivisions are done along the same axis. As I discovered, a large number of subdivisions may be realized as a series of thin bands (below). In this case, it might be best to subdivide larger regions over a series of calls, as I describe below.
Iterative Subparcellation
If banding among your subdivisions is a problem, here's an example of the iterative approach using region size, rather than a split file:
First, break down clusters into 400mm2 chunks:
mris_divide_parcellation fsaverage lh \ cache.th30.abs.sig.ocn.annot 400 400functional_subclusters
Next, break down the 400mm2 chunks into sub-200mm2 regions:
mris_divide_parcellation fsaverage lh \ 400functional_subclusters.annot \ 200 200functional_subclusters
Because each call to mris_divide_parcellation bisects the region along it's longest axis, successive function calls at each iteration produce increasingly less elongated subparcellations.
Transfer Subparcellation to Other Subjects
Once satisfied with the source subparcellation, the scheme can be mapped to other surfaces using mri_surf2surf if, for example, you are working in self space (this needs to be done individually for the lh and rh surfaces -- below is an example command for the left):
mri_surf2surf \ --srcsubject fsaverage \ --trgsubject FS_T1_501 \ --hemi lh \ --sval-annot $SUBJECTS_DIR/fsaverage/label/lh.200functional_subclusters.annot \ --tval $SUBJECTS_DIR/FS_T1_501/label/lh.200functional_subclusters.annot
Pro Tip: Use a global variable (e.g. SUBN) and use $SUBN in place of your subject in trgsubject and tval. Better yet, use the clustmap.sh shell script.
You may want to pull out overlapping voxels for T1 and T2 data (like a venn diagram) before mapping the surfaces to each subject, here.
Note: If you are working in fsaverage space for all participants, you only have to subparcellate your annotations once -- for the fsaverage surface. Because the functional analyses are invariably carried out in fsaverage space in order to compute group-level statistics, you will seldom have to transfer a subparcellation to a specific participant's surface space.