Mind Reading: Difference between revisions
| (51 intermediate revisions by 5 users not shown) | |||
| Line 3: | Line 3: | ||
Source data for input patterns are obtained using the same methods used for connectivity analyses: | Source data for input patterns are obtained using the same methods used for connectivity analyses: | ||
# Load surface-space time series data (<code>loadFSTS</code>) | # Load surface-space time series data (<code>loadFSTS</code>) | ||
# Remove any non-linear trends in the data (<code>NLDetrendFSTS</code>) | # Remove any non-linear trends in the data (<code>[[NLDetrendFSTS]]</code>) | ||
#*Note which, if any, rows you choose to drop from the data (e.g., it is customary to drop the first ~8-10 seconds of volumes from fMRI time series) | #*Note which, if any, rows you choose to drop from the data (e.g., it is customary to drop the first ~8-10 seconds of volumes from fMRI time series) | ||
# Normalize the time series data (<code>normalizeMatrix</code>) | # Normalize the time series data (<code>normalizeMatrix</code>) | ||
# Binarize and scale the normalized time series data (<code>binarizeMatrix</code>) | # Binarize and scale the normalized time series data (<code>[[binarizeMatrix]]</code>) | ||
== Determine Targets == | == Determine Targets == | ||
===Aggregating PsychToolBox Log Files=== | |||
A series of related .mat files produced by PsychToolBox can now be easily collected into a single cell array using the PTBParser.m MATLAB function, found in the ubfs/Scripts/Matlab folder. This function optionally takes a set of filenames, a path (default: current directory), and a vector of run numbers, which, if provided, will override the run numbers that are stored in the log files. This functionality is provided to avoid having multiple runs with the same number (e.g., if combining data from multiple sessions, each with runs 1:n). If multiple instances of the same run are given or are encoded in the log files, the runs are automatically renumbered 1:n in the order they appear in the filesystem (alphabetical, usually). | |||
eio=PTBParser(); %GUI selection box allows selection of multiple .mat files anywhere in filesystem | |||
eio=PTBParser('run', [1, 7, 2, 8, 3, 9, 4, 10, 5, 11, 6, 12]); | |||
%Selected files will be loaded in the order they appear in the filesystem | |||
%Log files 2, 4, 6, 8, 10 and 12 will be renumbered runs 7:12 | |||
%Log files 1, 3, 5, 7, 9, 11 will be renumbered runs 1:6 | |||
=== Classifying Task vs Baseline Blocks === | === Classifying Task vs Baseline Blocks === | ||
If the goal is simply to distinguish task block from rest periods, use <code>findBlockBoundaries</code> as follows: | If the goal is simply to distinguish task block from rest periods, use <code>findBlockBoundaries</code> as follows: | ||
| Line 58: | Line 67: | ||
schedule(firstvol:lastvol)=blockcondition; | schedule(firstvol:lastvol)=blockcondition; | ||
end | end | ||
=== Event-Related Classification === | |||
The previous example would be used to determine targets for a network to be trained on uninterrupted tasks within a block design. In the case of a mixed or event-related design, jitter may be incorporated in between events of the same condition, and/or events of different conditions may be intermixed. The approach below is to initialize a vector of targets for each volume with zeros (for fixation) and set the task condition targets only for volumes with a trial associated with that condition. | |||
==== TSTagger ==== | |||
A MATLAB function, [[TSTagger]] has been written to assign a label to a 5-second activity window from the fMRI time series. The first step is to marry the event information from PTBParser with the time series by creating a cell array of structs. This cell array is used to create a series of .csv files (1 for each time series) where the activity associated with the trials with the designated condition codes is tagged with the associated condition code. Activity values are calculated as the median activity within a 5-second window, offset by 1 second from the trial onset. For example, if an event happened at the 10 second mark, we calculate the median activity for each region within the 11- and 16-second mark time window. | |||
====="...But I only want events that meet specific criteria"===== | |||
No problem. We need to select the rows from the PTB Log file that we care about. In the example below, we're going to filter for trials with a modality code of 1 (VIS) or 2 (AUD). Assume we've already extracted the experiment info and stored it in dat.expinfo as in the above example. We simply iterate through the dat cell array of structs, find the column of interest (in this case, '''modality''') and find the rows that have the values of interest to make a filter. Then, replace the original expinfo.data with the filtered data structure. | |||
nruns=length(DAT); | |||
for i=1:nruns | |||
tmp=DAT{i}.expinfo.data; | |||
modalities=cell2mat(cellfun(@(x) double(x), {tmp.modality}, 'UniformOutput', false)); %This ugliness is because the script breaks if a run terminated early | |||
filter=(modalities=='''1'''|modalities=='''2'''); %which rows have the modalities of interest (1 or 2)? | |||
tmp=tmp(filter); %retain only the rows with the modalities of interest | |||
DAT{i}.expinfo.data=tmp; %replaces the original expinfo.data with the filtered data | |||
end | |||
==== csv File Cleanup ==== | |||
Some command-line cleanup to replace those condition codes with classifier unit index values will likely be required. In a shell terminal: | |||
SUB=0259 | |||
cat ${SUB}_*.csv > ${SUB}_hi.csv | |||
sed -i .bak 's/,11/,0/g' ${SUB}_hi.csv | |||
sed -i .bak 's/,21/,1/g' ${SUB}_hi.csv | |||
sed -i .bak 's/,31/,2/g' ${SUB}_hi.csv | |||
Of course, you may realize that it might be appropriate to differentiate the low-familiarity conditions for runs 1-6 (pre-exposure) from those for runs 7-12: | |||
cat ${SUB}_00[1-6].csv > ${SUB}_lo_pre.csv | |||
cat ${SUB}_00[7-9].csv ${SUB}_01[0-2].csv > ${SUB}_lo_post.csv | |||
sed -i .bak 's/,12/,0/g' ${SUB}_lo_pre.csv | |||
sed -i .bak 's/,22/,1/g' ${SUB}_lo_pre.csv | |||
sed -i .bak 's/,32/,2/g' ${SUB}_lo_pre.csv | |||
sed -i .bak 's/,12/,0/g' ${SUB}_lo_post.csv | |||
sed -i .bak 's/,22/,1/g' ${SUB}_lo_post.csv | |||
sed -i .bak 's/,32/,2/g' ${SUB}_lo_post.csv | |||
== Match Targets to Inputs == | == Match Targets to Inputs == | ||
Input values will come from time series data, imported and scaled as described [[Functional_Connectivity_(Neural_Network_Method) | here]]. If any volumes have been discarded from the input time series, the same volumes will need to be deleted from the schedule of targets. Assuming we have a schedule and a set of inputs for a single run, we need to match each input vector to the corresponding condition in the schedule vector and produce a MikeNet example file. This will use a Matlab function, tentatively called <code>mindReadingXFiles</code>. | Input values will come from time series data, imported and scaled as described [[Functional_Connectivity_(Neural_Network_Method) | here]]. If any volumes have been discarded from the input time series, the same volumes will need to be deleted from the schedule of targets. Assuming we have a schedule and a set of inputs for a single run, we need to match each input vector to the corresponding condition in the schedule vector and produce a MikeNet example file. This will use a Matlab function, tentatively called <code>mindReadingXFiles</code>. As usual, calling <code>help mindReadingXFiles</code> will provide you with some usage guidance. | ||
W=8; | |||
CTIMES=[0 1]; | |||
OTIMES=[2]; | |||
PREFIX='C2D3'; | |||
P=1; | |||
mindReadingXFiles('inputs', INPUTS, ... | |||
'targets', TASK_TARGETS, 'window', W, ... | |||
'clamp', CTIMES, 'output', OTIMES, ... | |||
'prolog', P, ... | |||
'prefix', PREFIX); | |||
This function will generate one or more ''.ex'' files with the specified filename prefix. A sliding window is used to generate the examples in each file. A window size of ''w'' (4 is the default) will present ''w'' consecutive input patterns, on successive time steps (e.g., when the window is set to 4, the first example will present the first, second, third and fourth activation patterns; the second example will present the second, third, fourth and fifth activation patterns, etc.). The target for each input pattern is the corresponding row from the TARGETS vector. The ''clamp'' parameter sets the time steps that each input pattern needs to be clamped for, and the ''output'' parameter sets the time offset from the start of an input pattern presentation to check the output activation. This pattern is continued until the window is filled (e.g., the first input pattern is clamped for 0, 1, with targets on 2; the second pattern is on 2, 3 with targets on 4; the third pattern is clamped on 4, 5 with targets on 6; etc.). The output offset should be selected to allow for inputs in a multilayer network to have an opportunity to influence the output layer. Additionally, a prolog value, ''p'' can be used to instruct the function to present the initial ''p'' input patterns without setting associated target values, and a 'nulltrain' value set to a value other than 0 will set null targets if no target values are associated with the pattern (generating an error value for any output nodes that are not turned off for these time points). | |||
=== Working with Null Events === | |||
If working from a set of targets that includes an explicit null event code (e.g., 0=fixation, 1=word, 2=pseudoword), it may be desirable to drop the implicit null events from the set of targets. Assuming the null events are assigned a code of 0, a function <code>noNulls</code>. Applied to a set of targets using <code>cellfun</code>, it sets all 0 targets to NaN, and then decrements by one the remaining codes (e.g., 1 becomes 0, 2 becomes 1, etc.): | |||
==== noNulls.m ==== | |||
function M=noNulls(A) | |||
M=A; | |||
M(A==0)=NaN; | |||
M=M-1; | |||
end | |||
==== Applying noNulls.m using cellfun ==== | |||
TASK_TARGETS=cellfun(@noNulls, TASK_TARGETS, 'UniformOutput', 0); | |||
%Note: the UniformOutput argument allows cellfun to return a cell array with differently-sized matrices | |||
== | == Graphing Continuous Performance == | ||
'''<span style="color:red;">Note:</span> this section only pertains to outputs of continuous networks, which you are likely not running. It's 99.9% likely that this section does not have information that is relevant to whatever it is you're working on.''' | |||
Load the network output into a matrix, '''''A'''''. The first two columns contain information about the trial and the time point; the remaining columns contain the output activations, and there should be one column for each classification condition. | Load the network output into a matrix, '''''A'''''. The first two columns contain information about the trial and the time point; the remaining columns contain the output activations, and there should be one column for each classification condition. | ||
%Isolate output activations | %Isolate output activations | ||
| Line 95: | Line 159: | ||
=== Plot Network Output Time Series and Overlay Event Onsets === | === Plot Network Output Time Series and Overlay Event Onsets === | ||
Finally, the <code>plot()</code> function will plot a vector of network outputs, and <code>vline()</code> or <code>gridxy()</code> is used to overlay a vertical line where each event occurred. | Finally, the <code>plot()</code> function will plot a vector of network outputs, and <code>vline()</code> or <code>gridxy()</code> is used to overlay a vertical line where each event occurred. If plotting a continuous time series, the <code>smooth()</code> function can be used to eliminate the spurious noise that occurs in the oversampled network time series. | ||
A=load('test.Early1.200000.gz.valid_Early_1.ex.txt'); | |||
V1=find(A(:,5)==1)-1; %subtracting because A(:,1) starts counting at 0 | |||
V2=find(A(:,6)==1)-1; %alternatively, could just add 1 to A(:,1) | |||
V3=find(A(:,7)==1)-1; | |||
figure(1); | figure(1); | ||
plot( | plot(A(:,1), A(:,2), 'r-*', A(:,1), A(:,3), 'g-*', A(:,1), A(:,4), 'b-*'); | ||
gridxy( | gridxy(V1, 'Color', 'r', 'Linestyle', ':'); | ||
gridxy(V2, 'Color', 'g', 'Linestyle', ':'); | |||
gridxy(V3, 'Color', 'b', 'Linestyle', ':'); | |||
gridxy( | |||
gridxy( | |||
== Project Specific Notes == | |||
# [[Deep Learning Semantic Category Classification]] | |||
[[Category: Time Series ]] | [[Category: Time Series ]] | ||
[[Category: Neural Networks ]] | [[Category: Neural Networks ]] | ||
[[Category: Machine Learning ]] | |||
Latest revision as of 20:32, 17 July 2019
The goal of so-called "mind reading" techniques is to use machine learning algorithms to decode brain states (as indexed by global patterns of cortical/subcortical activation) into their associated experimental conditions. Where this technique uses supervised learning, two things are required: First, a series of input patterns must be generated from fMRI activations. Second, a target pattern must be generated for each of the input patterns. This target pattern will indicate the experimental condition to be associated with each of the input patterns (e.g., REST (0) vs TASK (1)).
Determine Inputs
Source data for input patterns are obtained using the same methods used for connectivity analyses:
- Load surface-space time series data (
loadFSTS) - Remove any non-linear trends in the data (
NLDetrendFSTS)- Note which, if any, rows you choose to drop from the data (e.g., it is customary to drop the first ~8-10 seconds of volumes from fMRI time series)
- Normalize the time series data (
normalizeMatrix) - Binarize and scale the normalized time series data (
binarizeMatrix)
Determine Targets
Aggregating PsychToolBox Log Files
A series of related .mat files produced by PsychToolBox can now be easily collected into a single cell array using the PTBParser.m MATLAB function, found in the ubfs/Scripts/Matlab folder. This function optionally takes a set of filenames, a path (default: current directory), and a vector of run numbers, which, if provided, will override the run numbers that are stored in the log files. This functionality is provided to avoid having multiple runs with the same number (e.g., if combining data from multiple sessions, each with runs 1:n). If multiple instances of the same run are given or are encoded in the log files, the runs are automatically renumbered 1:n in the order they appear in the filesystem (alphabetical, usually).
eio=PTBParser(); %GUI selection box allows selection of multiple .mat files anywhere in filesystem
eio=PTBParser('run', [1, 7, 2, 8, 3, 9, 4, 10, 5, 11, 6, 12]);
%Selected files will be loaded in the order they appear in the filesystem
%Log files 2, 4, 6, 8, 10 and 12 will be renumbered runs 7:12
%Log files 1, 3, 5, 7, 9, 11 will be renumbered runs 1:6
Classifying Task vs Baseline Blocks
If the goal is simply to distinguish task block from rest periods, use findBlockBoundaries as follows:
%As mentioned above, keep in mind any samples dropped from the inputs: droprows=[1 2 3 4]; %this vector was used in the call to NLDetrendFSTS; sample_rate=2.047; %an fMRI study, with a TR=2.047 seconds b=findBlockBoundaries([], sample_rate); targets=zeros(1,b(end)); %1 zero for each volume -- default=baseline for block=1:size(b,1) targets(b(block,1):b(block,2))=1; %block volumes get a '1' end targets(droprows)=[]; %remove from the schedule the dropped time points %the first 4 × 2.047 seconds have been dropped from the schedule and from the input data %as if this period never happened
It is possible that the number of samples in the input data may exceed the number of targets. For example, the experiment script may terminate at the 6:00 mark, but the neuroimaging may continue for an additional few seconds, ending at, say the 6:04 mark. Any input samples extending beyond the timestamp associated with the final target value obtained using the above method would be undefined, though they could be construed as being associated with non-task events. To avoid ambiguity, however, it might be advisable to drop input samples that were obtained after the last target.
Classifying Task Blocks
If blocks are associated with different tasks or conditions to be classified, the schedule vector, schedule can be created similarly, but with some modification. Here's some examples.
sample_rate=2.047; %each sample spans 2.047 seconds in this fMRI example
expinfo=load('LDT_Sub_1004_Run_11_18-Apr-2016.mat');
t=cell2mat({expinfo.data.timestamp});
vols=floor(t/sample_rate)+1; %convert the timestamps into volume numbers
cond=double(cell2mat({expinfo.data.conditon})); %what condition is each trial?
%p.s., note the typo on "conditon"
b=double(cell2mat({expinfo.data.block})); %what block is each trial?
bnums=unique(b); %what are the different blocks?
lookup=[0,1;0,2]; %condition codes that make up each block condition
bcodes=[1,2];%code assigned to each block condition
schedule=zeros(1,vols(end));%blank schedule preallocated for each volume
%%This loop will iterate through all the numbered blocks and use the lookup
%%table to determine which block code to assign each block, depending on
%%the individual trial conditions present in that block.
%%Then, all individual volumes that belong to that block will get assigned
%%that condition code. Anything not assigned a block code will remain '0'
%%(or 'rest'/'baseline')
for i=1:length(bnums)
idx=find(b==bnums(i));%get indices of current block
codes=unique(cond(idx));%what conditions are represented in this block?
blockcondition=bcodes(ismember(lookup, codes, 'rows'));%lookup the blockcode (in bcodes) that matches the conditions in this block
firstvol=vols(idx(1));
lastvol=vols(idx(end));
schedule(firstvol:lastvol)=blockcondition;
end
Event-Related Classification
The previous example would be used to determine targets for a network to be trained on uninterrupted tasks within a block design. In the case of a mixed or event-related design, jitter may be incorporated in between events of the same condition, and/or events of different conditions may be intermixed. The approach below is to initialize a vector of targets for each volume with zeros (for fixation) and set the task condition targets only for volumes with a trial associated with that condition.
TSTagger
A MATLAB function, TSTagger has been written to assign a label to a 5-second activity window from the fMRI time series. The first step is to marry the event information from PTBParser with the time series by creating a cell array of structs. This cell array is used to create a series of .csv files (1 for each time series) where the activity associated with the trials with the designated condition codes is tagged with the associated condition code. Activity values are calculated as the median activity within a 5-second window, offset by 1 second from the trial onset. For example, if an event happened at the 10 second mark, we calculate the median activity for each region within the 11- and 16-second mark time window.
"...But I only want events that meet specific criteria"
No problem. We need to select the rows from the PTB Log file that we care about. In the example below, we're going to filter for trials with a modality code of 1 (VIS) or 2 (AUD). Assume we've already extracted the experiment info and stored it in dat.expinfo as in the above example. We simply iterate through the dat cell array of structs, find the column of interest (in this case, modality) and find the rows that have the values of interest to make a filter. Then, replace the original expinfo.data with the filtered data structure.
nruns=length(DAT);
for i=1:nruns
tmp=DAT{i}.expinfo.data;
modalities=cell2mat(cellfun(@(x) double(x), {tmp.modality}, 'UniformOutput', false)); %This ugliness is because the script breaks if a run terminated early
filter=(modalities==1|modalities==2); %which rows have the modalities of interest (1 or 2)?
tmp=tmp(filter); %retain only the rows with the modalities of interest
DAT{i}.expinfo.data=tmp; %replaces the original expinfo.data with the filtered data
end
csv File Cleanup
Some command-line cleanup to replace those condition codes with classifier unit index values will likely be required. In a shell terminal:
SUB=0259
cat ${SUB}_*.csv > ${SUB}_hi.csv
sed -i .bak 's/,11/,0/g' ${SUB}_hi.csv
sed -i .bak 's/,21/,1/g' ${SUB}_hi.csv
sed -i .bak 's/,31/,2/g' ${SUB}_hi.csv
Of course, you may realize that it might be appropriate to differentiate the low-familiarity conditions for runs 1-6 (pre-exposure) from those for runs 7-12:
cat ${SUB}_00[1-6].csv > ${SUB}_lo_pre.csv
cat ${SUB}_00[7-9].csv ${SUB}_01[0-2].csv > ${SUB}_lo_post.csv
sed -i .bak 's/,12/,0/g' ${SUB}_lo_pre.csv
sed -i .bak 's/,22/,1/g' ${SUB}_lo_pre.csv
sed -i .bak 's/,32/,2/g' ${SUB}_lo_pre.csv
sed -i .bak 's/,12/,0/g' ${SUB}_lo_post.csv
sed -i .bak 's/,22/,1/g' ${SUB}_lo_post.csv
sed -i .bak 's/,32/,2/g' ${SUB}_lo_post.csv
Match Targets to Inputs
Input values will come from time series data, imported and scaled as described here. If any volumes have been discarded from the input time series, the same volumes will need to be deleted from the schedule of targets. Assuming we have a schedule and a set of inputs for a single run, we need to match each input vector to the corresponding condition in the schedule vector and produce a MikeNet example file. This will use a Matlab function, tentatively called mindReadingXFiles. As usual, calling help mindReadingXFiles will provide you with some usage guidance.
W=8;
CTIMES=[0 1];
OTIMES=[2];
PREFIX='C2D3';
P=1;
mindReadingXFiles('inputs', INPUTS, ...
'targets', TASK_TARGETS, 'window', W, ...
'clamp', CTIMES, 'output', OTIMES, ...
'prolog', P, ...
'prefix', PREFIX);
This function will generate one or more .ex files with the specified filename prefix. A sliding window is used to generate the examples in each file. A window size of w (4 is the default) will present w consecutive input patterns, on successive time steps (e.g., when the window is set to 4, the first example will present the first, second, third and fourth activation patterns; the second example will present the second, third, fourth and fifth activation patterns, etc.). The target for each input pattern is the corresponding row from the TARGETS vector. The clamp parameter sets the time steps that each input pattern needs to be clamped for, and the output parameter sets the time offset from the start of an input pattern presentation to check the output activation. This pattern is continued until the window is filled (e.g., the first input pattern is clamped for 0, 1, with targets on 2; the second pattern is on 2, 3 with targets on 4; the third pattern is clamped on 4, 5 with targets on 6; etc.). The output offset should be selected to allow for inputs in a multilayer network to have an opportunity to influence the output layer. Additionally, a prolog value, p can be used to instruct the function to present the initial p input patterns without setting associated target values, and a 'nulltrain' value set to a value other than 0 will set null targets if no target values are associated with the pattern (generating an error value for any output nodes that are not turned off for these time points).
Working with Null Events
If working from a set of targets that includes an explicit null event code (e.g., 0=fixation, 1=word, 2=pseudoword), it may be desirable to drop the implicit null events from the set of targets. Assuming the null events are assigned a code of 0, a function noNulls. Applied to a set of targets using cellfun, it sets all 0 targets to NaN, and then decrements by one the remaining codes (e.g., 1 becomes 0, 2 becomes 1, etc.):
noNulls.m
function M=noNulls(A) M=A; M(A==0)=NaN; M=M-1; end
Applying noNulls.m using cellfun
TASK_TARGETS=cellfun(@noNulls, TASK_TARGETS, 'UniformOutput', 0); %Note: the UniformOutput argument allows cellfun to return a cell array with differently-sized matrices
Graphing Continuous Performance
Note: this section only pertains to outputs of continuous networks, which you are likely not running. It's 99.9% likely that this section does not have information that is relevant to whatever it is you're working on.
Load the network output into a matrix, A. The first two columns contain information about the trial and the time point; the remaining columns contain the output activations, and there should be one column for each classification condition.
%Isolate output activations Output=A(:,2:end);
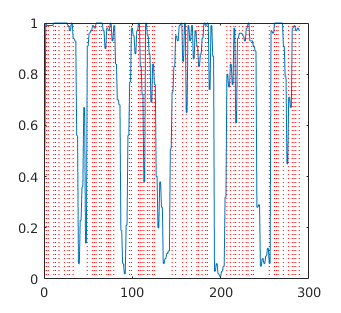
A visual indicator of network classification accuracy can be found by superimposing the volume onsets for each of the classification conditions. This can be done by using information from the schedule of TARGETS used to create the training file for the test data set (i.e., the matrix used by mindReadingXFiles.m) to create vertical indicator lines which are then superimposed on the network output plot. There are more than one MATLAB functions available on the MATLAB File Exchange. Two that I have tried are gridxy and vline.
Convert Schedule of Targets into Network Time Onsets
These steps are used to determine where each type of event occurred within the context of network time. Once you have the network timestamps, you can overlay the events on top of the network output time series. The example below shows how to determine where to plot the events associated with each volume in the test set
%%Obtain the events and timestamps associated with your test Exampleset
load('LDT_Sub_1004_Run_14_18-Apr-2016.mat'); %the data log for the run to be tested
sample_rate=2.047; %The sampling rate (TR) for this fMRI series
dropvols=4; %The first 4 samples were dropped from the time series data
t=cell2mat({expinfo.data.timestamp});
vols=floor(t/sample_rate)+1; %convert the event timestamps into volume numbers
vols=vols-dropvols; %Account for the initial dropped volumes
%%Convert sample timestamps into network time
%This will depend on the clamp/tdelay parameters used in the training/test example set
tstamps=(vols*2)-1; %Multiply the volumes by the clamp time and subtract 1 because the network is 0-indexed
conds=cell2mat({expinfo.data.conditon});%the vector of conditions associated with each event
onsets_0=tstamps(find(conds==0)); %when did the null events occur?
onsets_1=tstamps(find(conds==1)); %when did condition 1 occur?
onsets_2=tstamps(find(conds==2)); %when did condition 2 occur?
Plot Network Output Time Series and Overlay Event Onsets
Finally, the plot() function will plot a vector of network outputs, and vline() or gridxy() is used to overlay a vertical line where each event occurred. If plotting a continuous time series, the smooth() function can be used to eliminate the spurious noise that occurs in the oversampled network time series.
A=load('test.Early1.200000.gz.valid_Early_1.ex.txt');
V1=find(A(:,5)==1)-1; %subtracting because A(:,1) starts counting at 0
V2=find(A(:,6)==1)-1; %alternatively, could just add 1 to A(:,1)
V3=find(A(:,7)==1)-1;
figure(1);
plot(A(:,1), A(:,2), 'r-*', A(:,1), A(:,3), 'g-*', A(:,1), A(:,4), 'b-*');
gridxy(V1, 'Color', 'r', 'Linestyle', ':');
gridxy(V2, 'Color', 'g', 'Linestyle', ':');
gridxy(V3, 'Color', 'b', 'Linestyle', ':');